How to structure the manifest metadata file
Background
SODA helps you prepare the manifest metadata files conveniently. We explain here how it must be structured according to the SPARC rules in order to provide some insight about the structure of the file generated by SODA.
How to
- Format: the manifest file is accepted in either xlsx, csv, or json format. SODA generates it in the xlsx format based on the template provided by the Curation Team. A screenshot of a sample manifest file from a primary folder is provided below:
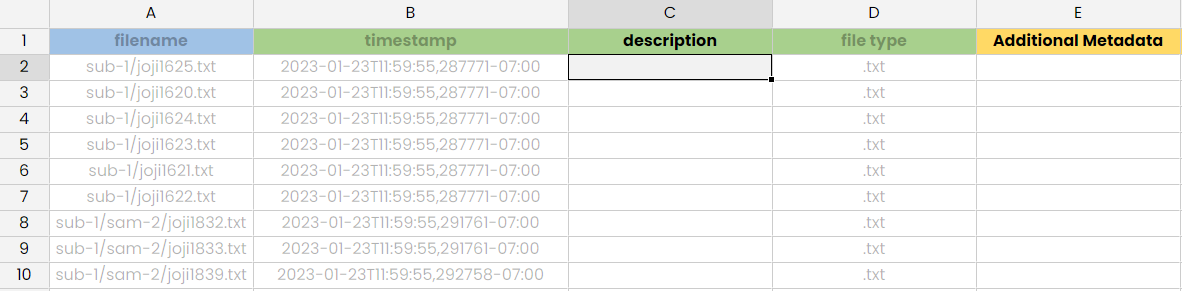
- Location in the dataset: a manifest file is required in either each of the high-level SPARC folders OR in each of the dataset folders with at least one file.
- SODA follows the structure #1 when generating the manifest files automatically for you. We thus only talk about structure #1 below. Should be
Noneif no extension (instead of blank). - Content: The
filenameandtimestampfields are mandatory and it is suggested to provide other fields as needed to understand each file.- Filename: This should be path to the files relative to the high-level SPARC folder where the manifest file is located (with a forward slash independently of the OS). Note that the extension of the file is included.
- Timestamp: The last time the file was modified locally on the creator's computer, preferably in ISO 8601 format with UTC offset. For example, this would be
2019-07-13T02:43:14+05:30for a file last modified on July 13th 2019 at 2:43 AM (14 seconds) in PST (thus +5:30 to show the offset from UTC). If this timestamp is not available, the Pennsieve's timestamp (which indicate the last time the file was modified on Pennsieve) could be used as an alternative. - File type: Type of this file. Would be
.jpgorImage filefor the file histology.jpg.